gapseq
Informed prediction and analysis of bacterial metabolic pathways and genome-scale networks
Purpose
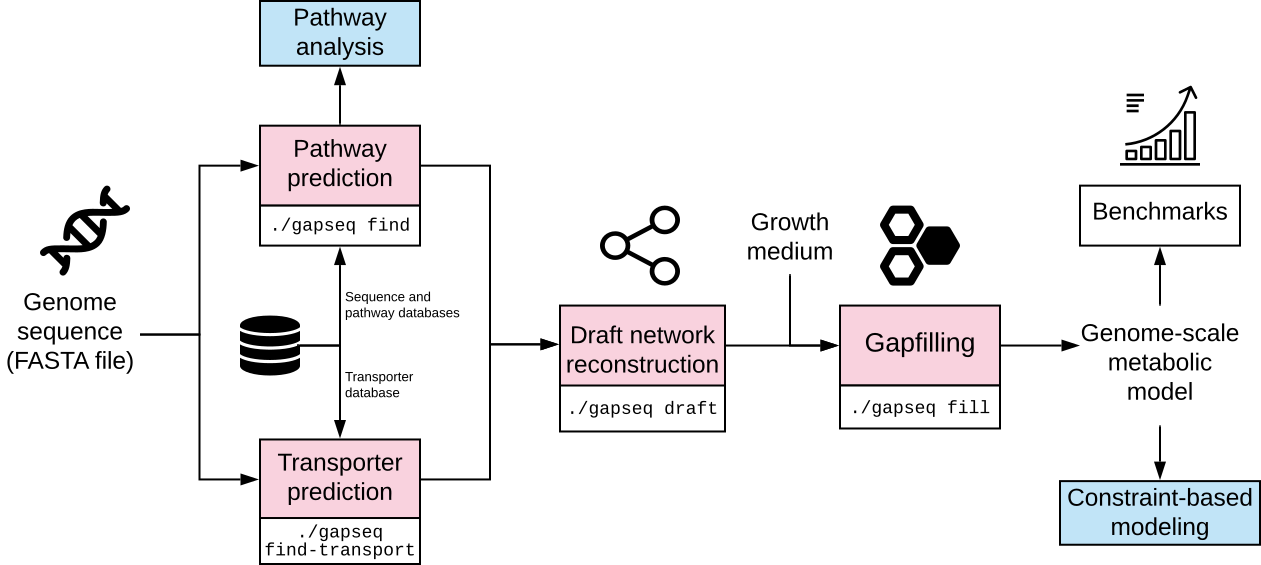
gapseq is designed to combine metabolic pathway analysis with metabolic network reconstruction and curation. Based on genomic information and databases for pathways and reactions, gapseq can be used for:
- prediction of metabolic pathways from various databases
- transporter inference
- metabolic model construction (genome-scale metabolic networks)
- multi-step gap filling
Links
Citing gapseq
Zimmermann J, Kaleta C, Waschina S.
gapseq: informed prediction of bacterial metabolic pathways and reconstruction of accurate metabolic models
Genome Biology. 2021. 22(1). doi: 10.1186/s13059-021-02295-1
Eutropia
Agent-based metabolic modelling of microbial communities in time and continuous µ-meter-scale space
Purpose
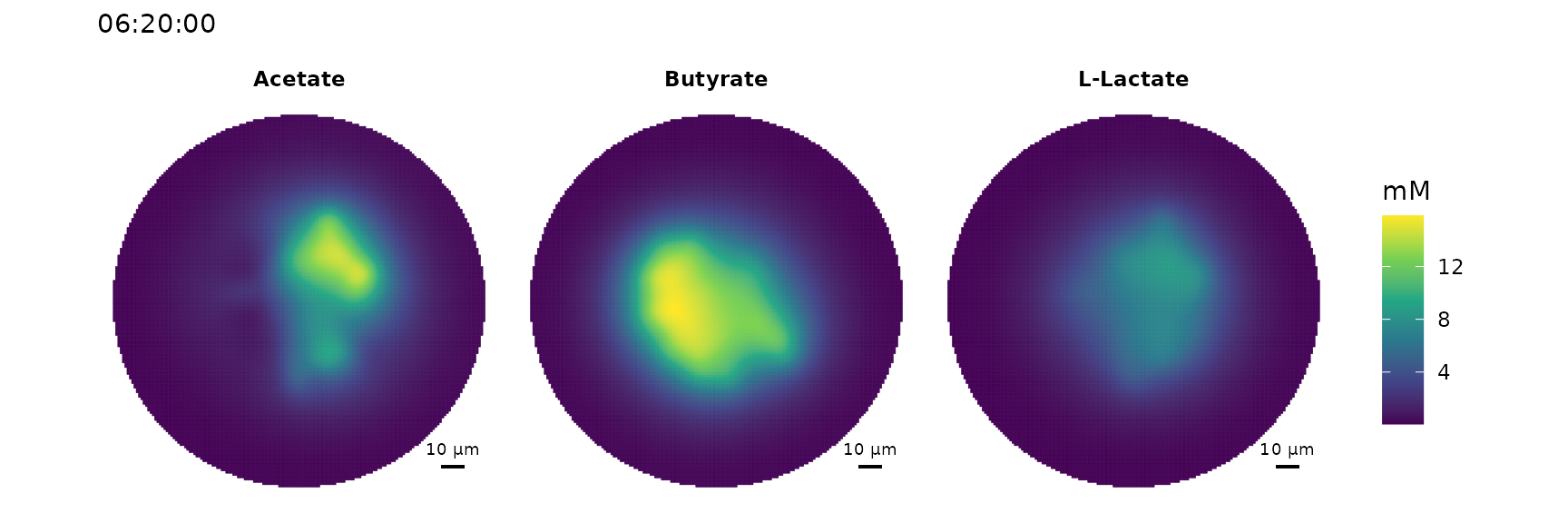
Eutropia is a R-package for cell agent-based metabolic modelling of microbial communities. It allows dynamic simulations of two-dimensional surface-attached cell communities. A few features of Eutropia, that you might find interesting:
- Complex polygons (also non-convex) as growth environment
- Extracellular enzymes
- Nutrient regimes
- Chemotaxis (attracting and repelling)
- Direct calculations of community assortment and segregation (see e.g. Yanni et al. (2015) Current Biology)
Links
 Nutriinformatics
Nutriinformatics
